Iceberg, Parquet and query engines
2025-04-25
During my time at Huawei, one of the biggest problem we had was "how to store our observability data". Our initial database was based on OpenGemini, but we were always looking for places to improve.
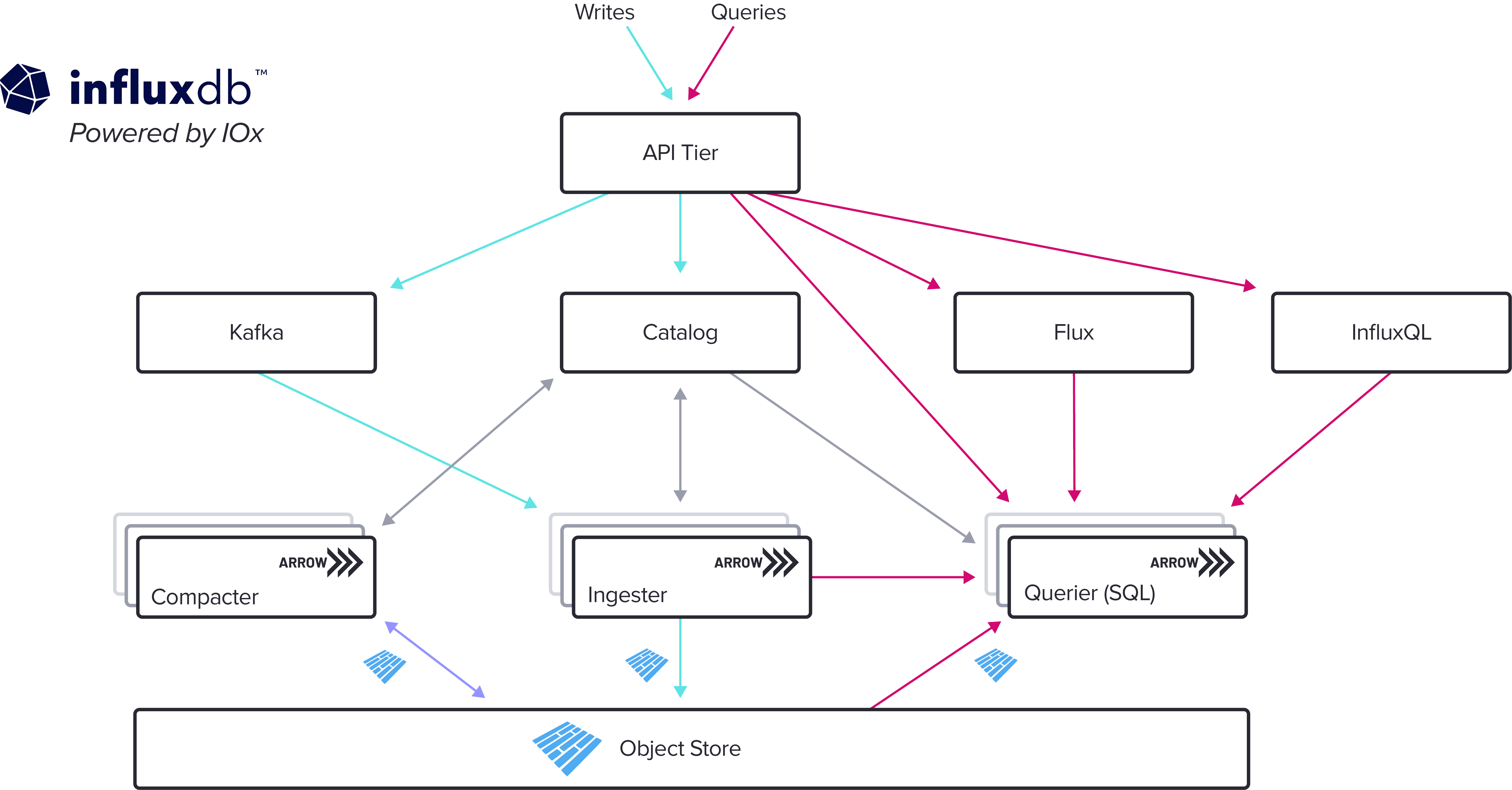
A common design for this type of database is to split the compute nodes from the storage, a very recent example of this is the new InfluxDB written in Rust (ref). The data is stored in a Object Store (such as S3), while the other nodes handle the ingestion and querying (query execution plan, physical and logical).
Another example is ScopeDB (ref), which is very similar:
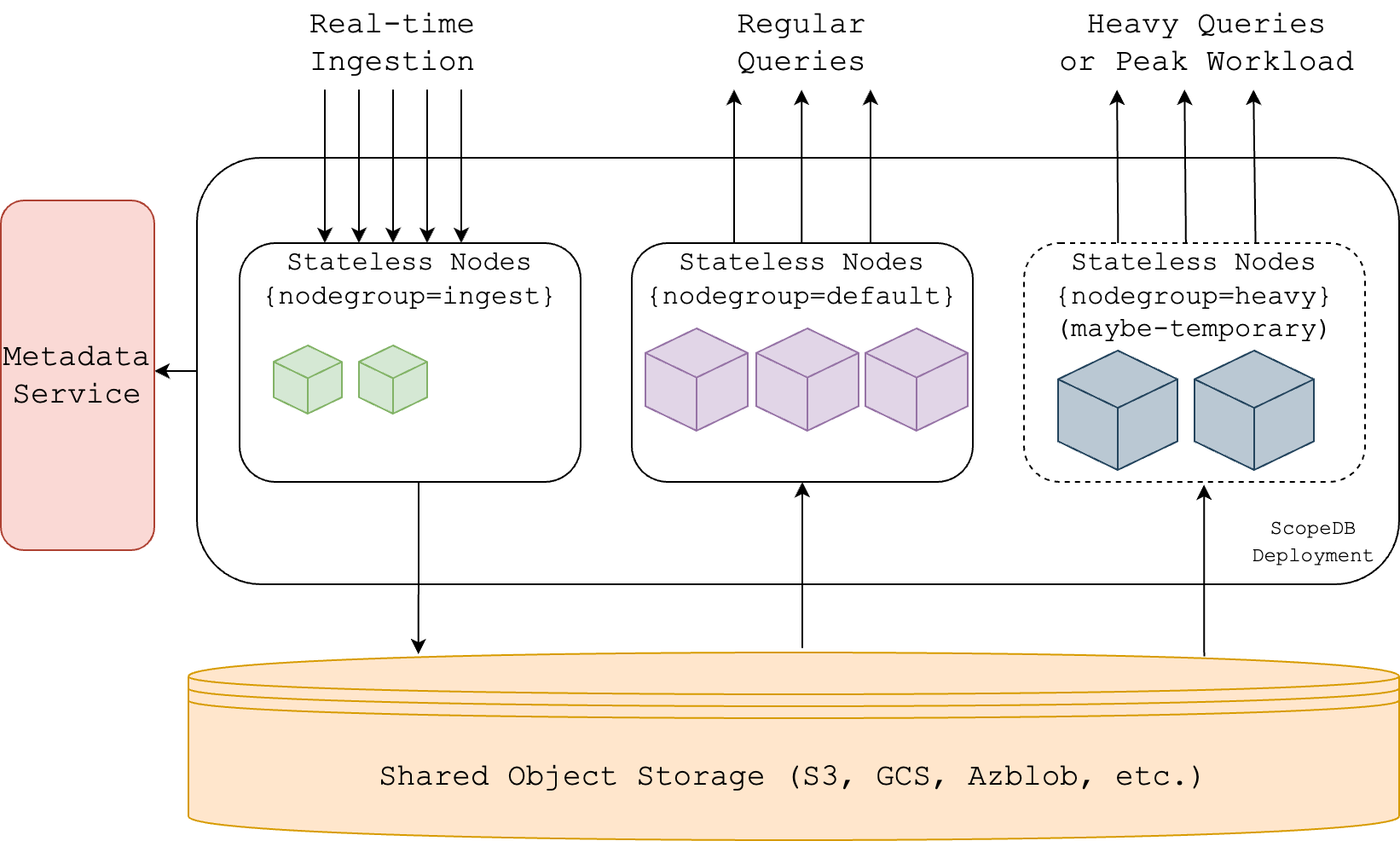
Since I took both images from the blogs of both companies, their terms are not normalized. InfludDB has no "Metadata Service", while ScopeDB has no "Catalog". Why? They seem so similar, they sure need similar components, right?
Well, a catalog is the Metadata Service, it works as a "structural blueprint", storing metadata and organizing schemas.
Anyway, going back to those databases. Within the Big Data scene, we have a bunch of new products build on top of the same standards:
- Parquet: Parquet is a columnar storage format. It's a self describing format, if you read the file from disk, you can find out the schema of the data by reading its metadata. Another very cool thing, is that Parquet keeps a summary of each column of the file, such as min, max and distinct_count.
- Arrow: While Parquet describes the format stored in disk, Arrow describes how the data should be kept in memory. It's a columnar format, perfect for data interchange and in-memory analytics.
- Iceberg: Iceberg is a table format. The idea is to manage a large, slow-changing collection of files in a distributed file system or key-value store as a table. Basically, you can save different Parquet files in S3 and use Iceberg to manage them (insert, delete, select, ...).
If you add your compute nodes on top of those three standards, you can create a toy-database. You basically need a node to handle ingestion. Those nodes should write to the Object Storage and update the Iceberg metadata with information about it. Using icerberg-rs and arrow-rs, this seems more or less like:
/// In this example, let's assume we will receive the data somehow as Arrow RecordBatch.
async fn write(&self, name: TableIdent, batch: RecordBatch) -> Result<(), Error> {
// iceberg transaction
let table = self.catalog.load_table(&name).await?;
let transaction = Transaction::new(&table);
let mut fast_append = transaction.fast_append(None, vec![])?;
// metadata for parquet writer
let file_io = table.file_io();
let location = DefaultLocationGenerator::new(table.metadata().clone())?;
// writing files to parquet first
let prefix = format!("{}-{}", name.name, Uuid::new_v4());
let file_name_generator = DefaultFileNameGenerator::new(
prefix,
None, //suffix
iceberg::spec::DataFileFormat::Parquet, //format
);
let parquet_props = parquet::file::properties::WriterProperties::builder().build();
let parquet_writer_builder = ParquetWriterBuilder::new(
parquet_props,
table.metadata().current_schema().clone(),
file_io.clone(),
location,
file_name_generator,
);
// just one partition `0` for now
let data_file_writer_builder = DataFileWriterBuilder::new(parquet_writer_builder, None, 0);
let mut writer = data_file_writer_builder.build().await?;
writer.write(batch).await?;
let data_files = writer.close().await?;
// now write to the metadata of our table
fast_append.add_data_files(data_files)?;
let transaction = fast_append.apply().await?;
transaction.commit(&*self.catalog).await?;
Ok(())
}
The query engine is the hardest part here, let's assume you are going to use something like like sqlparser to handle the parsing of SQL. You need to create a plan for how to fetch the data the user wants and how to execute it. For a toy project, you can push down most predicates and projections down to Iceberg. Remember that Parquet keeps some summary data about the columns? Iceberg does the same with its metadata, so it can decide if you need to open that file and do a scan there or skip it. Once you have your Parquet files, you can load them into memory using Arrow format and handle everything else you haven't pushdown to Iceberg.
This part is so big, that is hard to even show a small useful code here, but I am currently playing with this idea in a public repo which you can check it out. I'm far from done (as I write this text), so bare with me as I improve that code :).